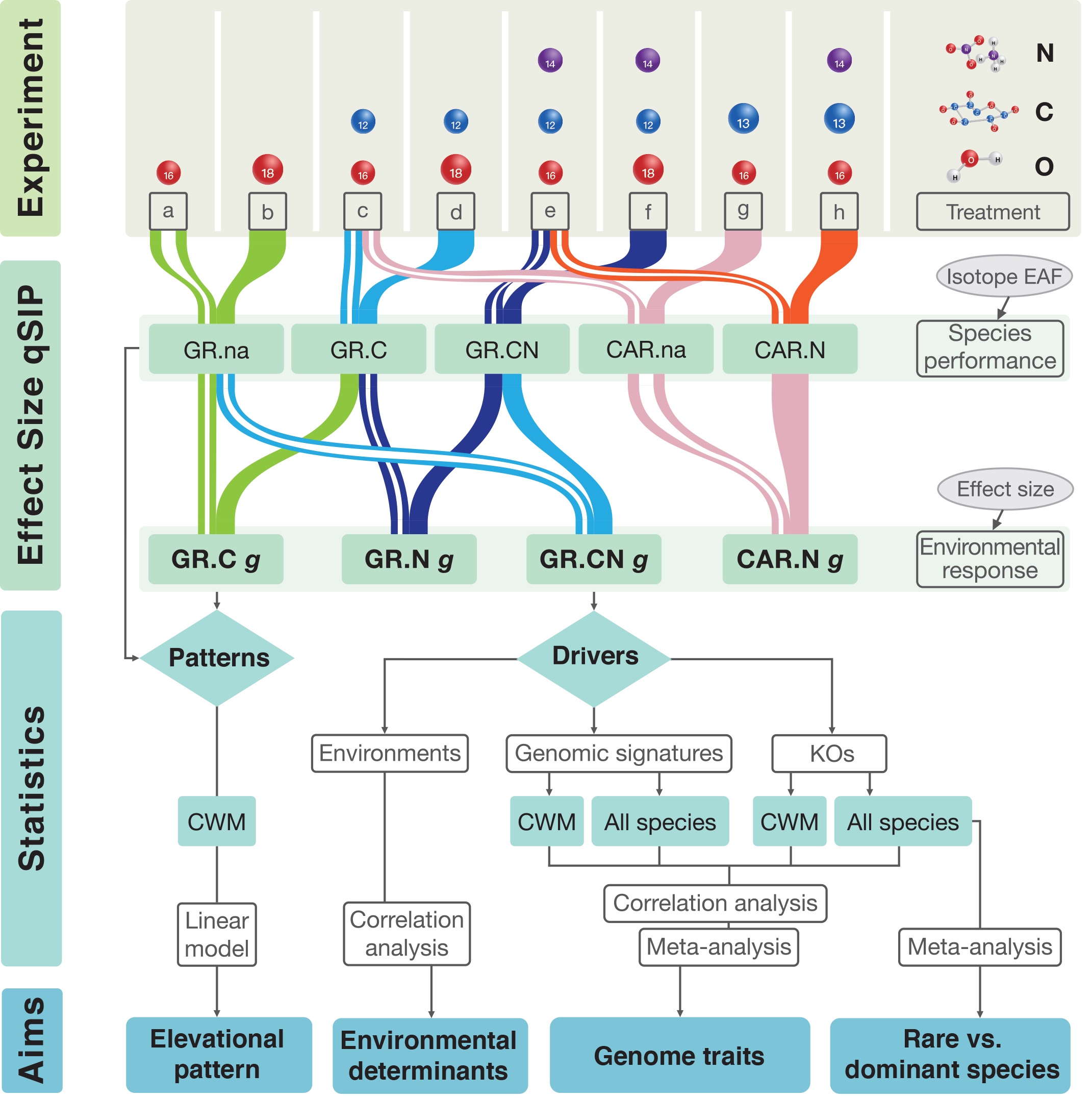
How does microbial species performance respond to environmental changes? Such a simple question is challenging to answer with quantitative measurements, especially for complex communities. We just got a paper published on line in Ecology to help in answering this question.
Based on stable isotope labelling of DNA, we propose a new approach namely effect-size qSIP to quantify the species environmental responses of microbes by comparing the species performance between defined control and treatment groups. We further tested this approach with unprecedented qSIP experimental data on mountainsides from Morrissey et al (2019 Nat Ecol Evol 3:1064-1069) and found that rare and dominant species differentially respond to nutrient enrichment via their metabolic traits.
Here comes the ABSTRACT:
How microbial species performance indicators, such as growth rate and carbon assimilation rate, respond to environmental changes is a challenging question, especially for complex communities. This limits our ability to understand how species performance responses to environmental changes (that is, species environmental responses) of microbes could be linked to genomic traits and nutrient availability. Based on stable isotope labelling of DNA, we propose a new approach with effect size metrics to quantify the species environmental responses of microbes by comparing the species performance between defined control and treatment groups. The species performance within microbial communities of the natural or altered environments could be quantitatively determined with quantitative stable isotope probing (qSIP). We further apply this approach namely effect size qSIP to measure species environmental responses upon carbon and nitrogen additions for soil bacteria on mountainsides and to understand their responses from the perspective of genomic traits. Towards high elevations, there is a stronger nitrogen limitation which is indicated by the higher aggregated responses, measured as community-weighted means, of bacterial growth rate upon nitrogen additions. The aggregated responses are further explained by genomic traits, which show higher percentages of significant Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologues (KOs) and more diverse KEGG pathways under nutrient additions including nitrogen and further improve the explanatory power of microbial environmental responses. Nitrogen-induced responses at the species level show the strongest associations with essential KOs for rare species, while carbon for dominant species. We conclude that, in addition to environmental determinants such as nitrogen limitation, genomic traits are extremely important for predicting microbial environmental responses at both the community and species levels. Taking advantage of this new approach at the species level, we reveal that rare and dominant species differentially respond to nutrient enrichment via their metabolic traits. The approach and findings can lead to a more holistic understanding of microbial environmental responses in natural habitats, which will be essential for predicting microbial community responses to global environmental changes.
Reference:
A. Hu, M. Ren, J. Wang. Microbial species performance responses to environmental changes: genomic traits and nutrient availability. Ecology, 2021. DOI:10.1002/ecy.3382 [Abstract][FullText]