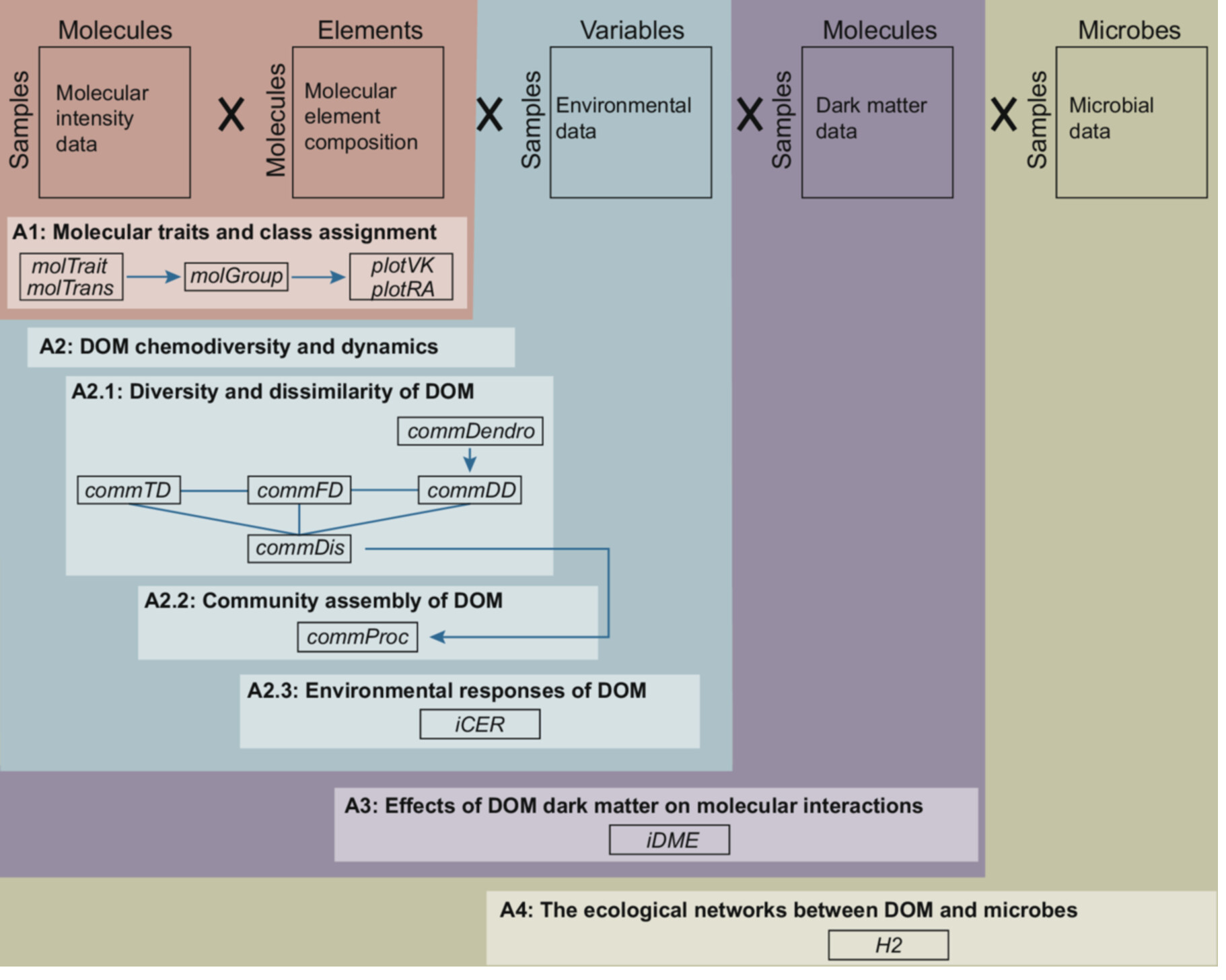
We recently developed a R packge iDOM for DOM statistical analyses, which is now published on line by mLife.
The study introduces iDOM, a new R package designed to analyze and visualize dissolved organic matter (DOM) data from FT-ICR MS. It supports various DOM data types, integrates environmental and microbial data, and includes tools for both basic and advanced statistical analyses. iDOM helps researchers explore DOM’s ecological roles, particularly under global changes like climate warming. By bridging chemistry and ecology, it offers a comprehensive framework for interpreting DOM’s complexity in ecosystems. The package and example datasets are available at GitHub.
Here is a introduction of this paper:
Abstract
Dissolved organic matter (DOM) contains thousands of molecules and is key for biogeochemical cycles in aquatic and terrestrial ecosystems by interacting with microbes. Over the last decade, the study of DOM has been advanced and accelerated with the developments of instrumental and statistical approaches. However, it is still challenging in statistical analyses, data visualization, and theoretical interpretations largely due to the complexity of molecular composition and underlying ecological mechanisms. In this study, we developed an R package iDOM with functions for the basic and advanced statistical analyses and the visualization of DOM derived from Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS). The package could handle various data types of DOM, including molecular compositional data, molecular traits, and uncharacterized molecules (i.e., dark matter). It could integrate explanatory data, such as environmental and microbial data, to explore the relationships between DOM and abiotic or biotic drivers. To illustrate its use, we presented case studies with an example dataset of DOM and microbial communities under experimental warming. We included case studies of basic functions for the calculation of molecular traits, the assignment of molecular classes, and the compositional analyses of chemical diversity and dissimilarity. We further showed the case studies with advanced functions to quantify DOM assembly processes, assess the effects of dark matter on molecular interactions, analyze the ecological networks between DOM and microbes, and explore their response to warming. The source code and example dataset of iDOM are publicly available on https://github.com/jianjunwang/iDOM. We expect that iDOM will serve as a comprehensive pipeline for DOM statistical analyses and bridge the gap between chemical characterization and ecological interpretation in a theoretical framework.
Impact statement
Dissolved organic matter (DOM) is a key component of Earth's ecosystems, playing a vital role in nutrient cycling and interactions with microbial communities. However, studying DOM presents significant statistical challenges due to its complex molecular composition and the difficulty of connecting its composition to ecological processes. To address these challenges, we developed iDOM, a new R package designed to streamline the analysis, visualization, and interpretation of DOM data. By integrating DOM data with environmental and microbial datasets, iDOM enables researchers to better understand how DOM responds to global changes, such as climate warming. This tool bridges disciplines of chemistry and ecology, providing novel insights into the DOM's role in ecosystem functioning.
How to cite
Meng F, Hu A, Jang K-S, Wang J. iDOM: Statistical analysis of dissolved organic matter characterized by high-resolution mass spectrometry. mLife. 2025; 1–13. https://doi.org/10.1002/mlf2.70002